JAXPM: A JAX-Based Framework for Scalable and Differentiable Particle Mesh Simulations
Wassim Kabalan
Alexandre Boucaud, François Lanusse
2025-05-01


Outline for This Presentation
- Beyond Summary Statistics Inference in Cosmology
- Building N-body Simulators for Cosmological Inference
- Modeling Observables: Weak Lensing & Lightcones
- Scaling Up: Distributed, Differentiable Simulations
The Traditional Approach to Cosmological Inference
- cosmological parameters (Ω): matter density, dark energy, etc.
- Predict observables: CMB, galaxies, lensing
- Extract summary statistics: \(P(k)\), \(C_\ell\) , 2PCF
- Compute likelihood: \(L(\Omega \vert data)\)
- Estimate \(\hat{\Omega}\) via maximization (\(\chi^2\) fitting)
Summary Statistics Based Inference
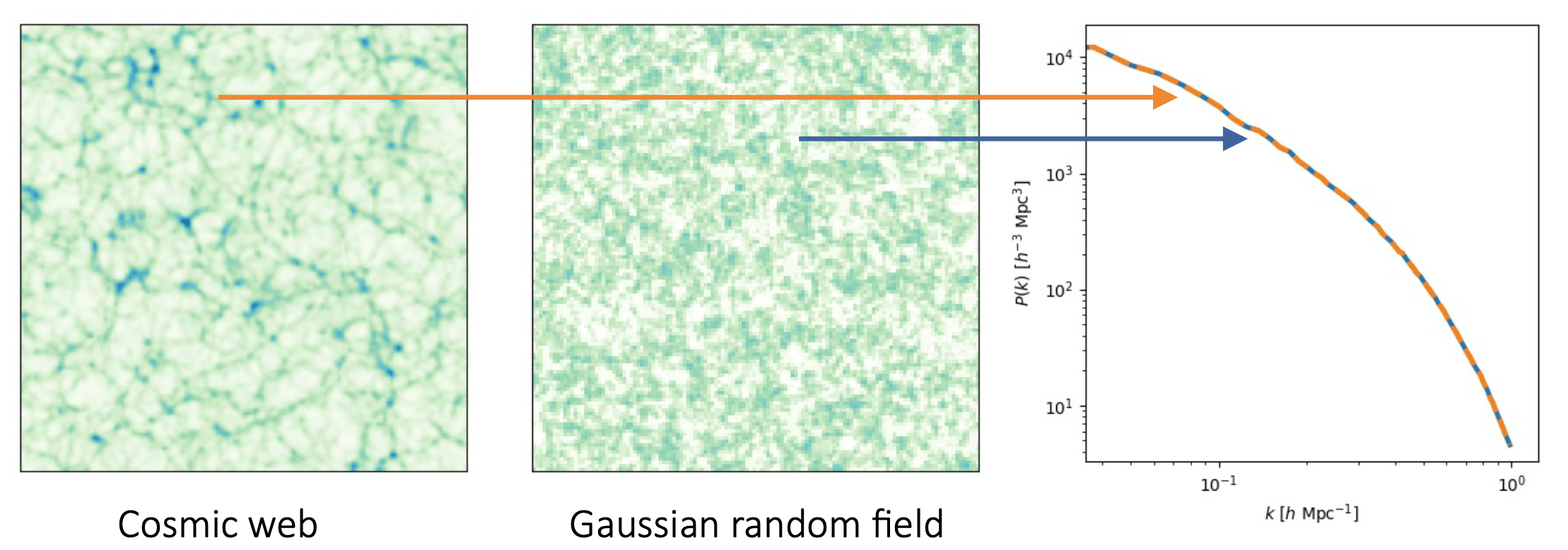
- Traditional inference uses summary statistics to compress data.
- Power spectrum fitting: \(P(k)\), \(C_\ell\)
- It misses complex, non-linear structure in the data
The Traditional Approach to Cosmological Inference
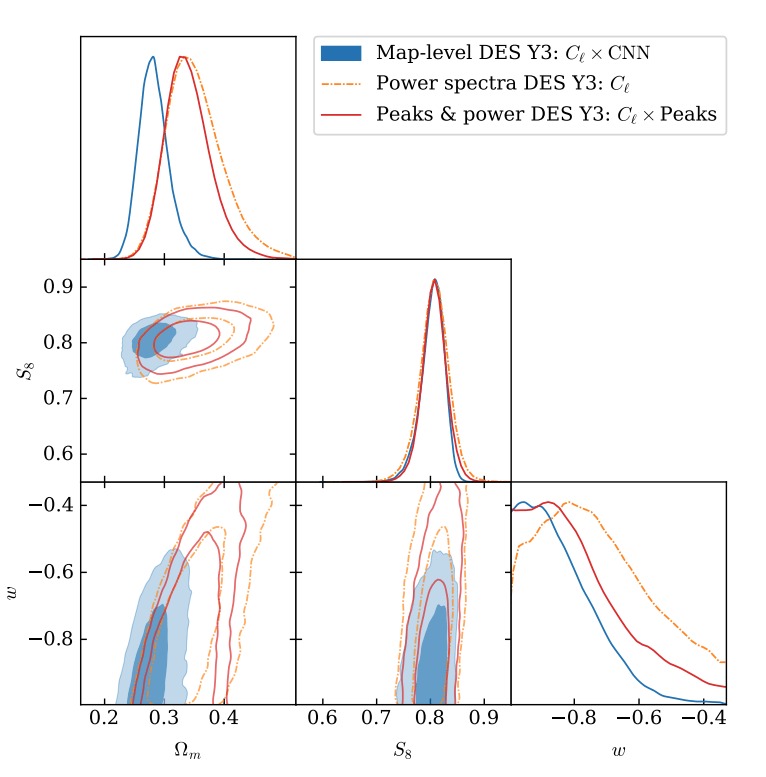
- Summary statistics (e.g. P(k)) discard the non-Gaussian features.
- Gradient-based curve fitting does not recover the true posterior shape.
How to maximize the information gain?
From Summary Statistics to Likelihood Free Inference
Bayes’ Theorem
\[ p(\theta \mid x_0) \propto p(x_0 \mid \theta) \cdot p(\theta) \]
- Prior: Encodes our assumptions about parameters \(\theta\)
- Likelihood: How likely the data \(x_0\) is given \(\theta\)
- Posterior: What we want to learn — how data updates our belief about \(\theta\)
- Simulators become the bridge between cosmological parameters and observables.
- How to use simulators allow us to go beyond summary statistics?
From Summary Statistics to Likelihood Free Inference
Implicit Inference
Treats the simulator as a black box — we only require the ability to simulate \((\theta, x)\) pairs.
No need for an explicit likelihood — instead, use simulation-based inference (SBI) techniques
Often relies on compression to summary statistics \(t = f_\phi(x)\), then approximates \(p(\theta \mid t)\).
Explicit Inference
Requires a differentiable forward model or simulator.
Treat the simulator as a probabilistic model and perform inference over the joint posterior \(p(x \mid \theta, z)\)
Computationally demanding — but provides exact control over the statistical model.
Implicit inference
Simulation-Based Inference Loop
- Sample parameters \(\theta_i \sim p(\theta)\)
- Run simulator \(x_i = p(x \vert \theta_i)\)
- Compress observables \(t_i = f_\phi(x_i)\)
- Train a density estimator \(\hat{p}_\Phi(\theta \mid f_\phi(x))\)
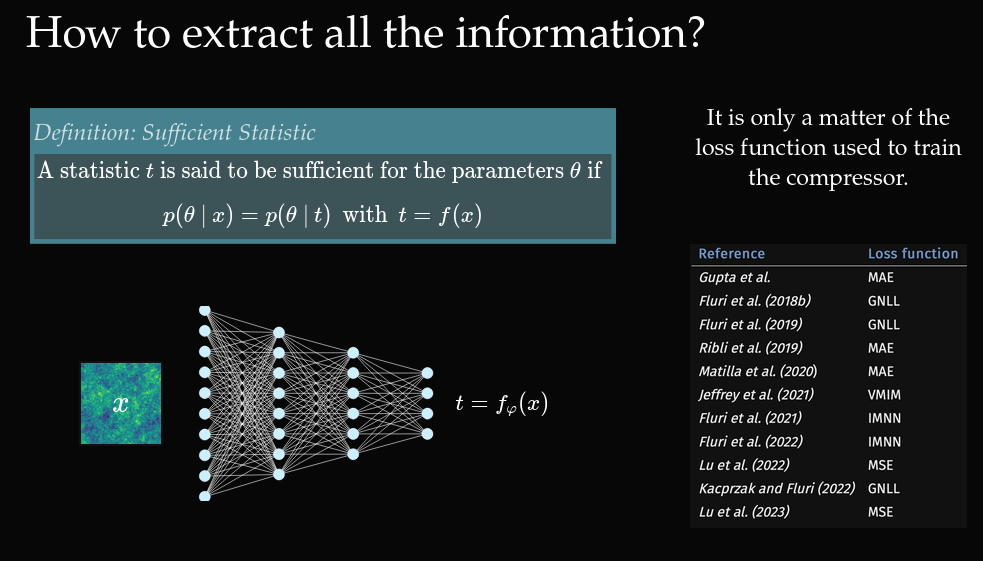
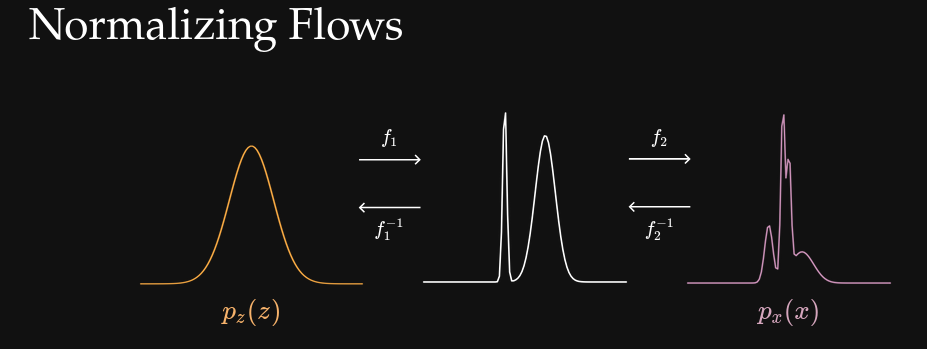
- Neural Summarisation (Zeghal & Lanzieri et al 2025).
- Normalizing Flows (Zeghal et al. 2022).
- ✅ Works with non-differentiable or stochastic simulators
- ❌ Requires an optimal compression function \(f_\phi\)
Explicit inference
The goal is to reconstruct the entire latent structure of the Universe — not just compress it into summary statistics. To do this, we jointly infer:
\[ p(\theta, z \mid x) \propto p(x \mid \theta, z) \, p(z \mid \theta) \, p(\theta) \]
Where:
\(\theta\): cosmological parameters (e.g. matter density, dark energy, etc.)
\(z\): latent fields (e.g. initial conditions of the density field)
\(x\): observed data (e.g. convergence maps or galaxy fields)
The challenge of explicit inference
The latent variables \(z\) typically live in very high-dimensional spaces — with millions of degrees of freedom.
Sampling in this space is intractable using traditional inference techniques.
- We need samplers that can scale efficiently to high-dimensional latent spaces and Exploit gradients from differentiable simulators
- This makes differentiable simulators essential for modern cosmological inference.
- Particle Mesh (PM) simulations offer a scalable and differentiable solution.
Particle Mesh Simulations
Compute Forces via PM method
- Start with particles \(\mathbf{x}_i, \mathbf{p}_i\)
- Interpolate to mesh: \(\rho(\mathbf{x})\)
- Solve Poisson’s Equation: \[ \nabla^2 \phi = -4\pi G \rho \]
- In Fourier space: \[ \mathbf{f}(\mathbf{k}) = i\mathbf{k}k^{-2}\rho(\mathbf{k}) \]
Time Evolution via ODE
- PM uses Kick-Drift-Kick (symplectic) scheme:
- Drift: \(\mathbf{x} \leftarrow \mathbf{x} + \Delta a \cdot \mathbf{v}\)
- Kick: \(\mathbf{v} \leftarrow \mathbf{v} + \Delta a \cdot \nabla \phi\)
- Fast and scalable approximation to gravity.
- A cycle of FFTs and interpolations.
- Sacrifices small-scale accuracy for speed and differentiability.
- Current implementations JAXPM v0.1, PMWD and BORG.
Using Full-Field Inference with Weak Lensing
From 3D Structure to Lensing Observables
- Simulate structure formation over time, taking snapshots at key redshifts
- Stitch these snapshots into a lightcone, mimicking the observer’s view of the universe
- Combine contributions from all slabs to form convergence maps
- Use the Born approximation to simplify the lensing calculation
Born Approximation for Convergence
\[ \kappa(\boldsymbol{\theta}) = \int_0^{r_s} dr \, W(r, r_s) \, \delta(\boldsymbol{\theta}, r) \]
Where the lensing weight is:
\[ W(r, r_s) = \frac{3}{2} \, \Omega_m \, \left( \frac{H_0}{c} \right)^2 \, \frac{r}{a(r)} \left(1 - \frac{r}{r_s} \right) \]
Can we start doing inference?
The impact of resolution on simulation accuracy
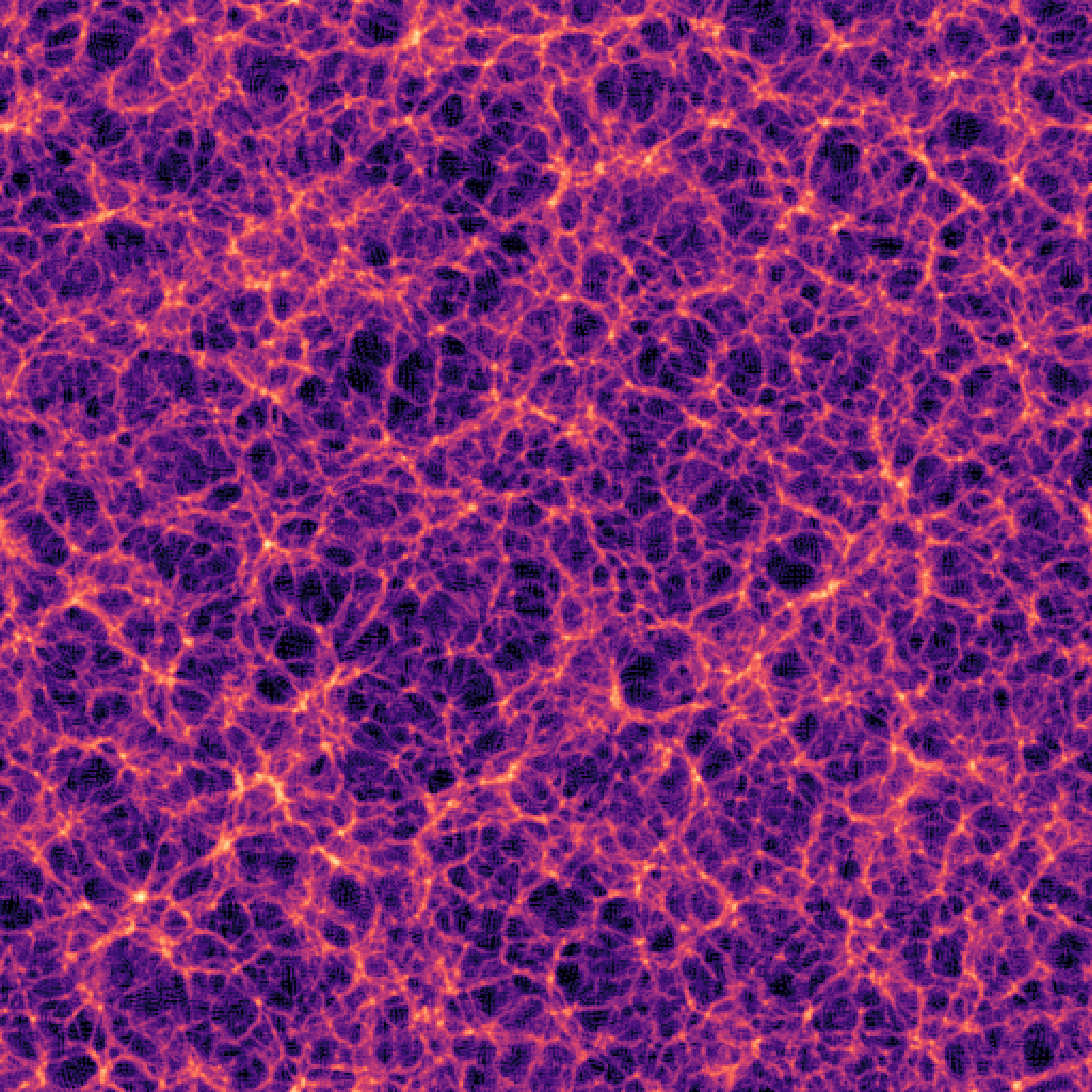
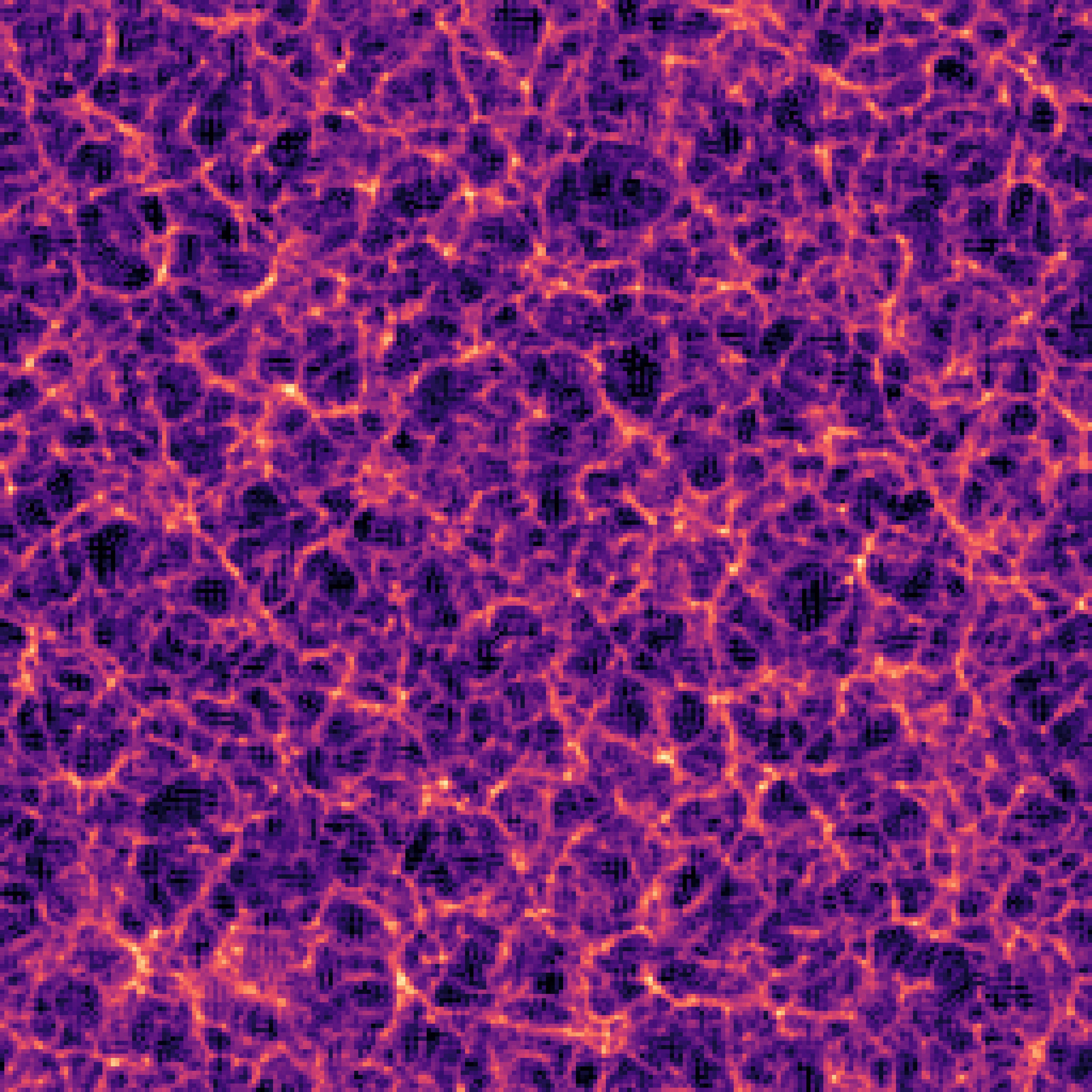
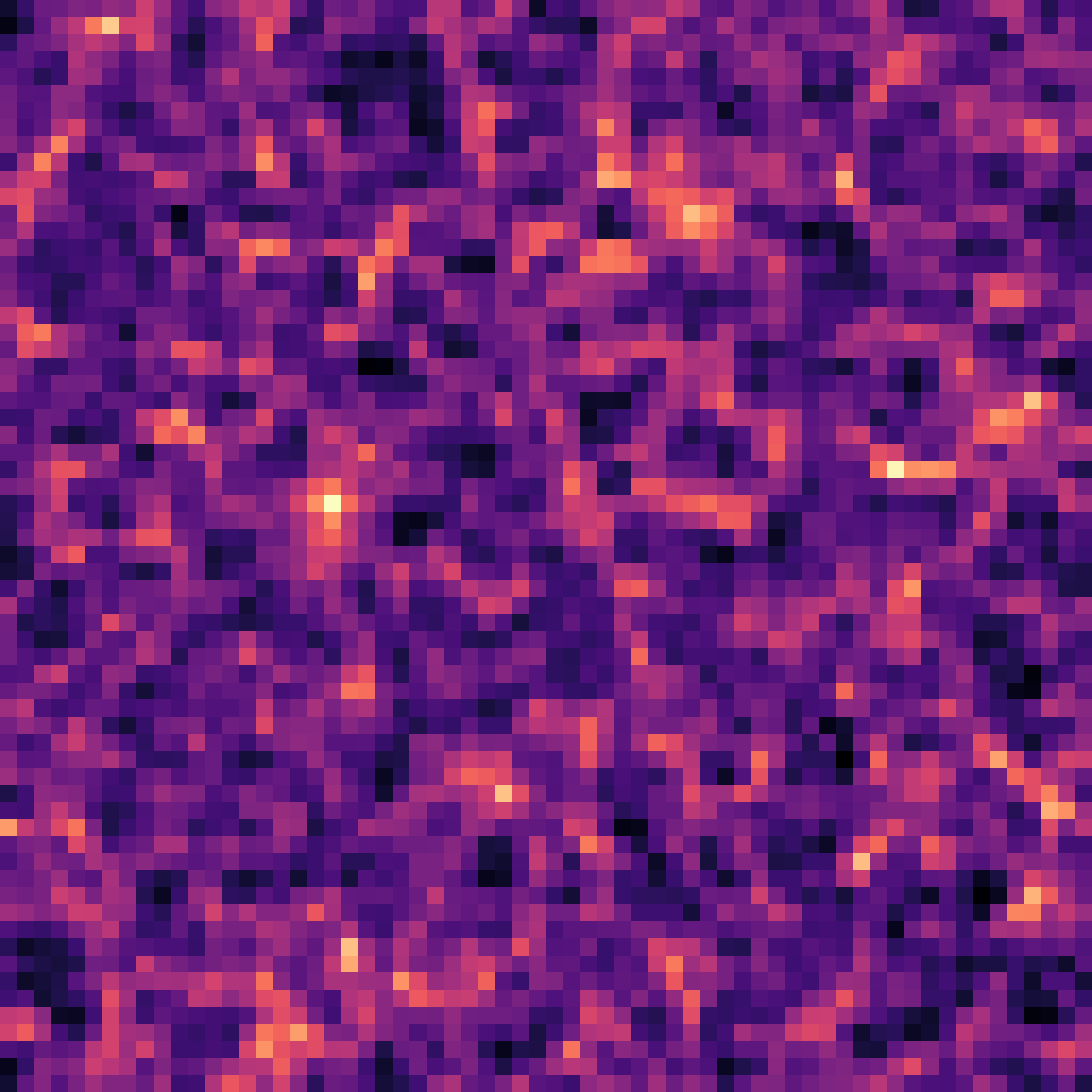
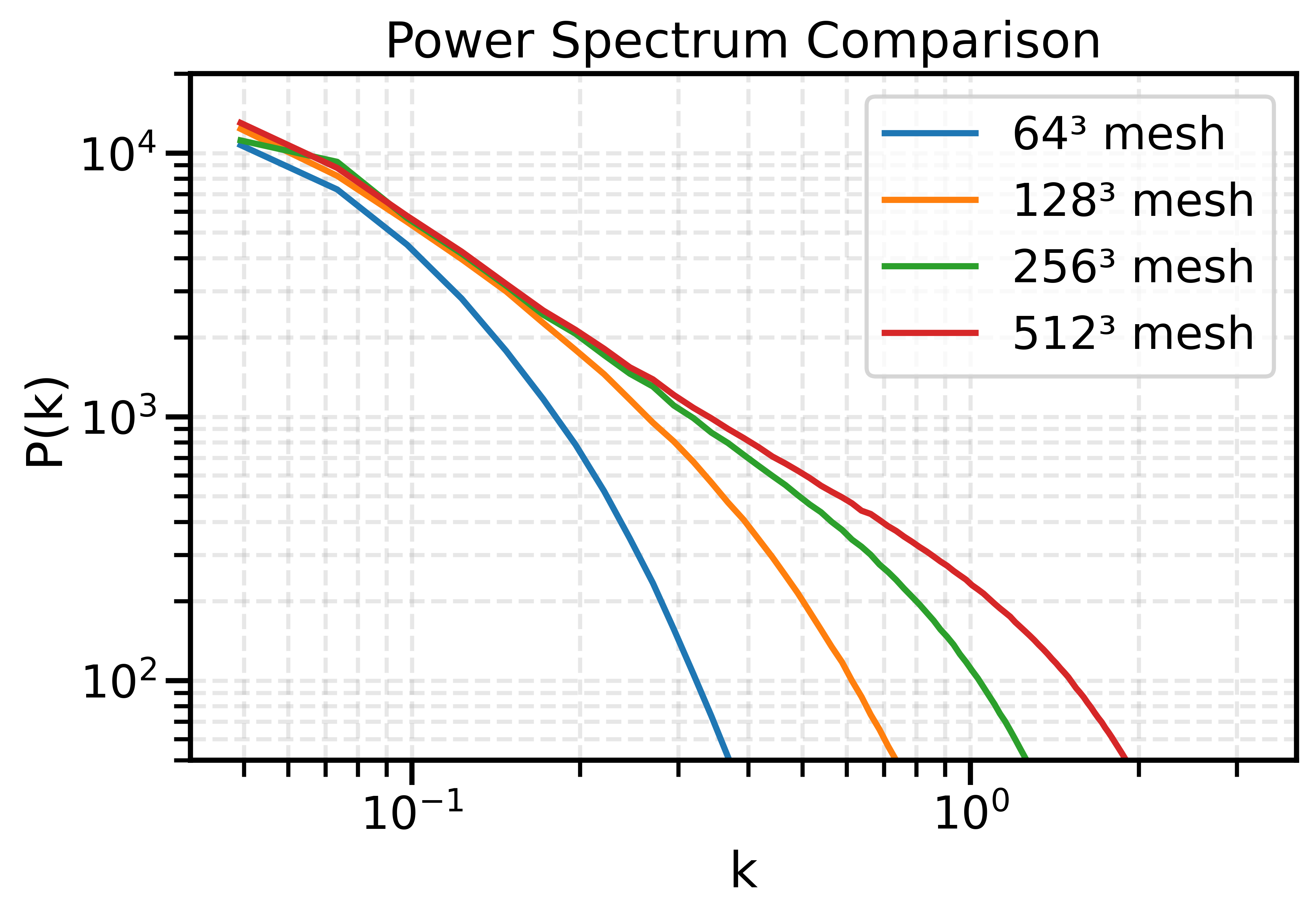
When the Simulator Fails, the Model Fails
Inference is only as good as the simulator it depends on.
- If we want to model complex phenomena like galaxy painting, baryonic feedback, or non-linear structure formation, our simulator must be not only fast, but also physically accurate.
- A decent resolution for weak lensing requires about 1 grid cell per Mpc/h — both for angular resolution and structure fidelity.
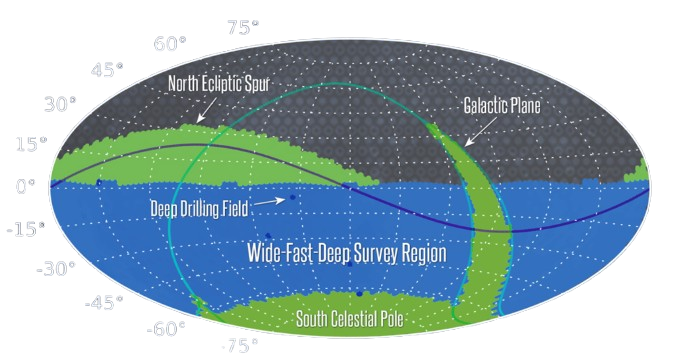
Scaling Up the simulation volume: The LSST Challenge
LSST Scan Range
- Covers ~18,000 deg² (~44% of the sky)
- Redshift reach: up to z ≈ 3
- Arcminute-scale resolution
- Requires simulations spanning thousands of Mpc in depth and width
- Simulating even a (1 Gpc/h)³ subvolume at 1024³ mesh resolution requires:
- ~54 GB of memory for a simulation with a single snapshot
- Gradient-based inference and multi-step evolution push that beyond 100–200 GB
Takeaway
- LSST-scale cosmological inference demands multiple (Gpc/h)³ simulations at high resolution.
Modern high-end GPUs cap at ~80 GB, so even a single box requires multi-GPU distributed simulation — both for memory and compute scalability.

We Need Scalable Distributed Simulations
Distributed Particle Mesh Simulation
Force Computation is Easy to Parallelize
Poisson’s equation in Fourier space:
\[ \nabla^2 \phi = -4\pi G \rho \]Gravitational force in Fourier space:
\[ \mathbf{f}(\mathbf{k}) = i\mathbf{k}k^{-2}\rho(\mathbf{k}) \]Each Fourier mode \(\mathbf{k}\) can be computed independently using JAX
Perfect for large-scale, parallel GPU execution

Fourier Transform requires global communication
jaxDecomp: Distributed 3D FFT and Halo Exchange
- Distributed 3D FFT using domain decomposition
- Fully differentiable, runs on multi-GPU and multi-node setups
- Designed as a drop-in replacement for
jax.numpy.fft.fftn
- Open source and available on PyPI \(\Rightarrow\)
pip install jaxdecomp - Halo exchange for mass conservation across subdomains
Distributed Particle Mesh Simulation
Cloud In Cell (CIC) Interpolation

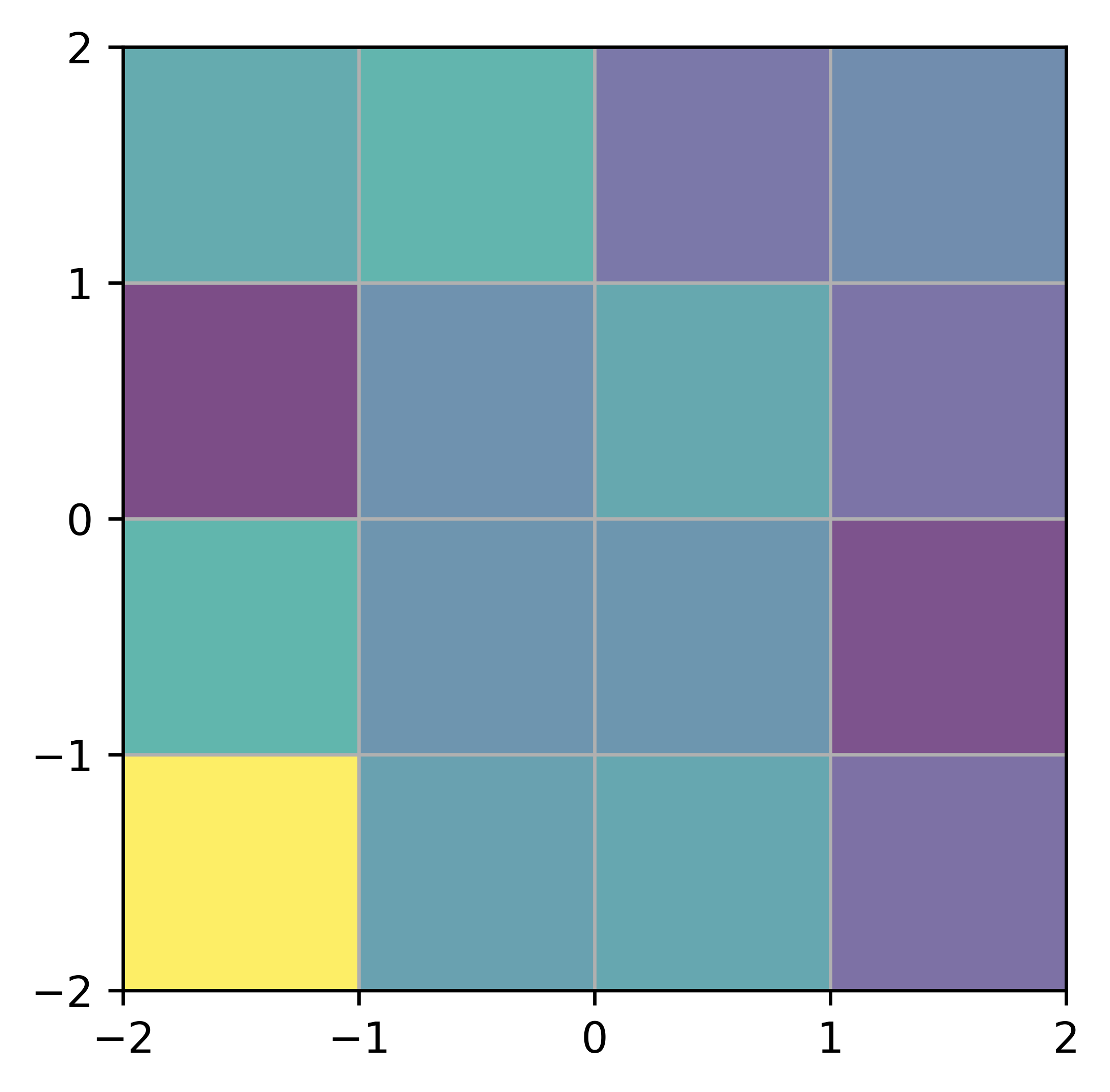
Mass Assignment and Readout
The Cloud-In-Cell (CIC) scheme spreads particle mass to nearby grid points using a linear kernel:
- Paint to Grid (mass deposition): \[ g(\mathbf{j}) = \sum_i m_i \prod_{d=1}^{D} \left(1 - \left|p_i^d - j_d\right|\right) \]
- Read from Grid (force interpolation): \[ v_i = \sum_{\mathbf{j}} g(\mathbf{j}) \prod_{d=1}^{D} \left(1 - \left|p_i^d - j_d\right|\right) \]
Distributed Cloud In Cell (CIC) Interpolation
In distributed simulations, each subdomain handles a portion of the global domain
Boundary conditions are crucial to ensure physical continuity across subdomain edges
CIC interpolation assigns and reads mass from nearby grid cells — potentially crossing subdomain boundaries
To avoid discontinuities or mass loss, we apply halo exchange:
- Subdomains share overlapping edge data with neighbors
- Ensures correct mass assignment and gradient flow across boundaries
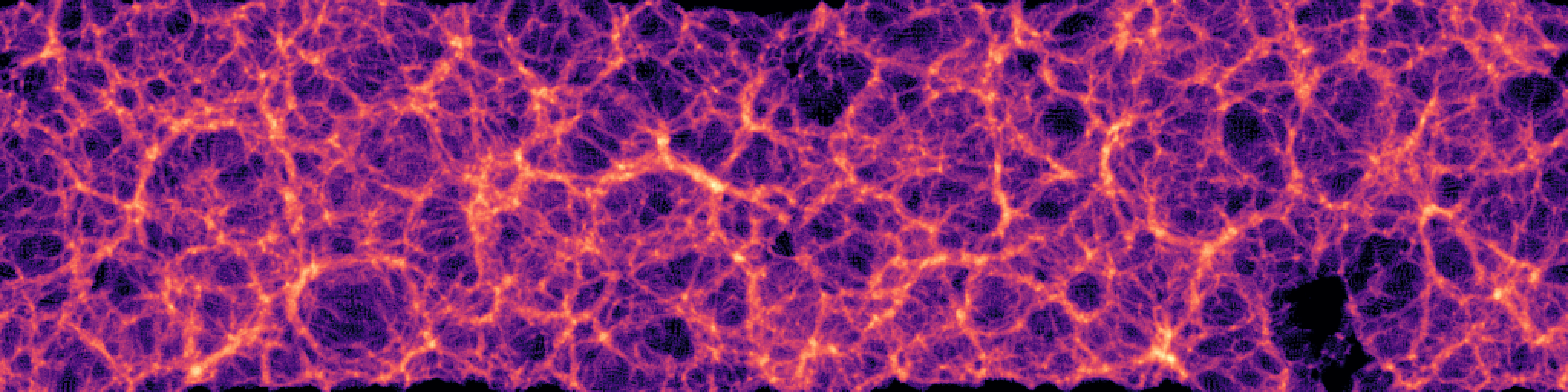
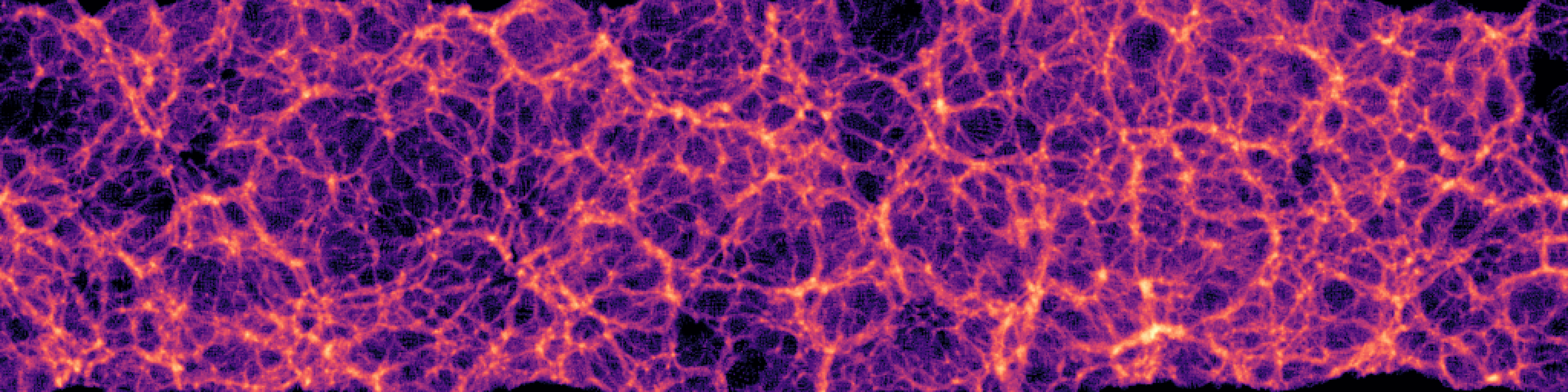
Without Halo Exchange (Not Distributed)
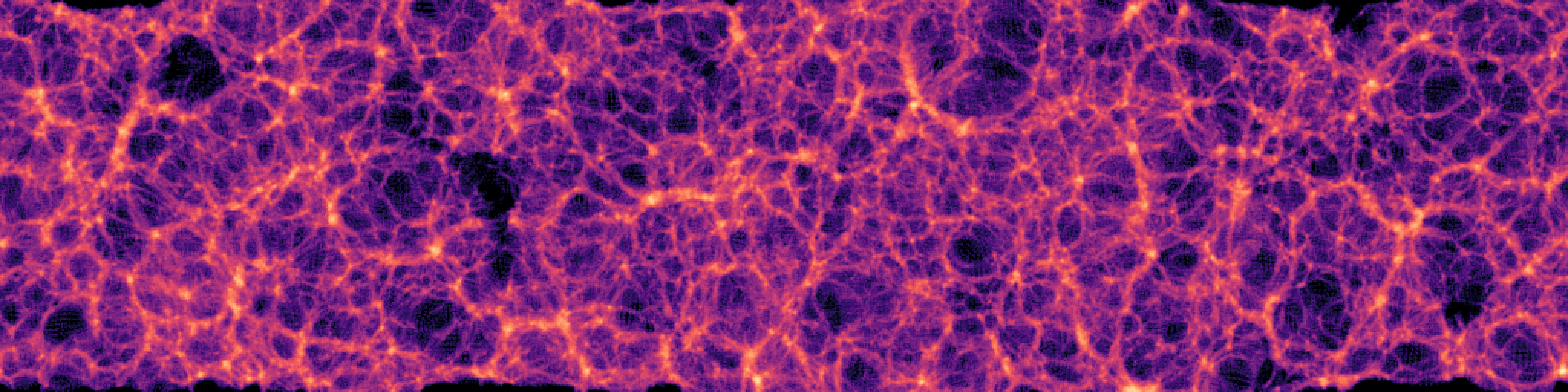
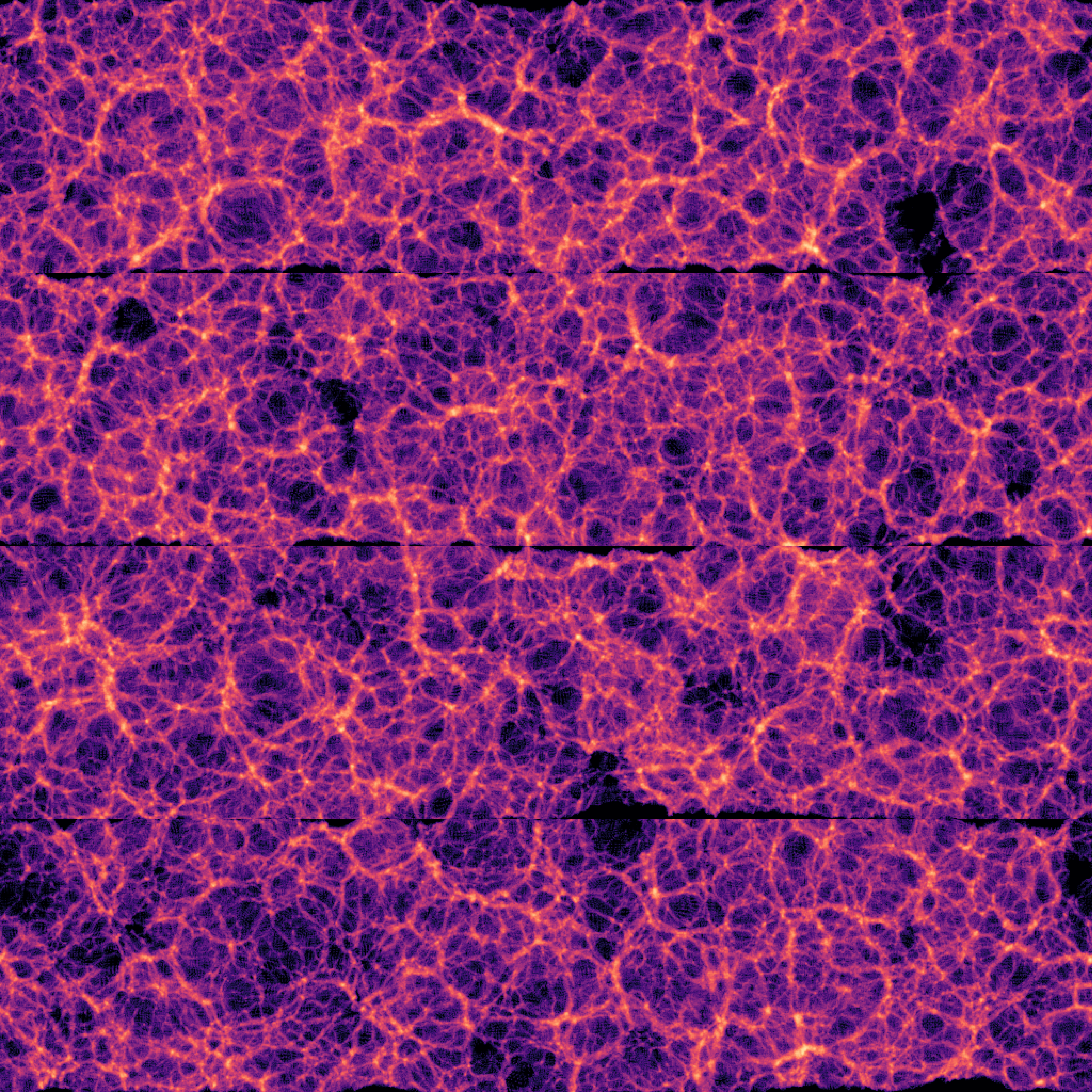
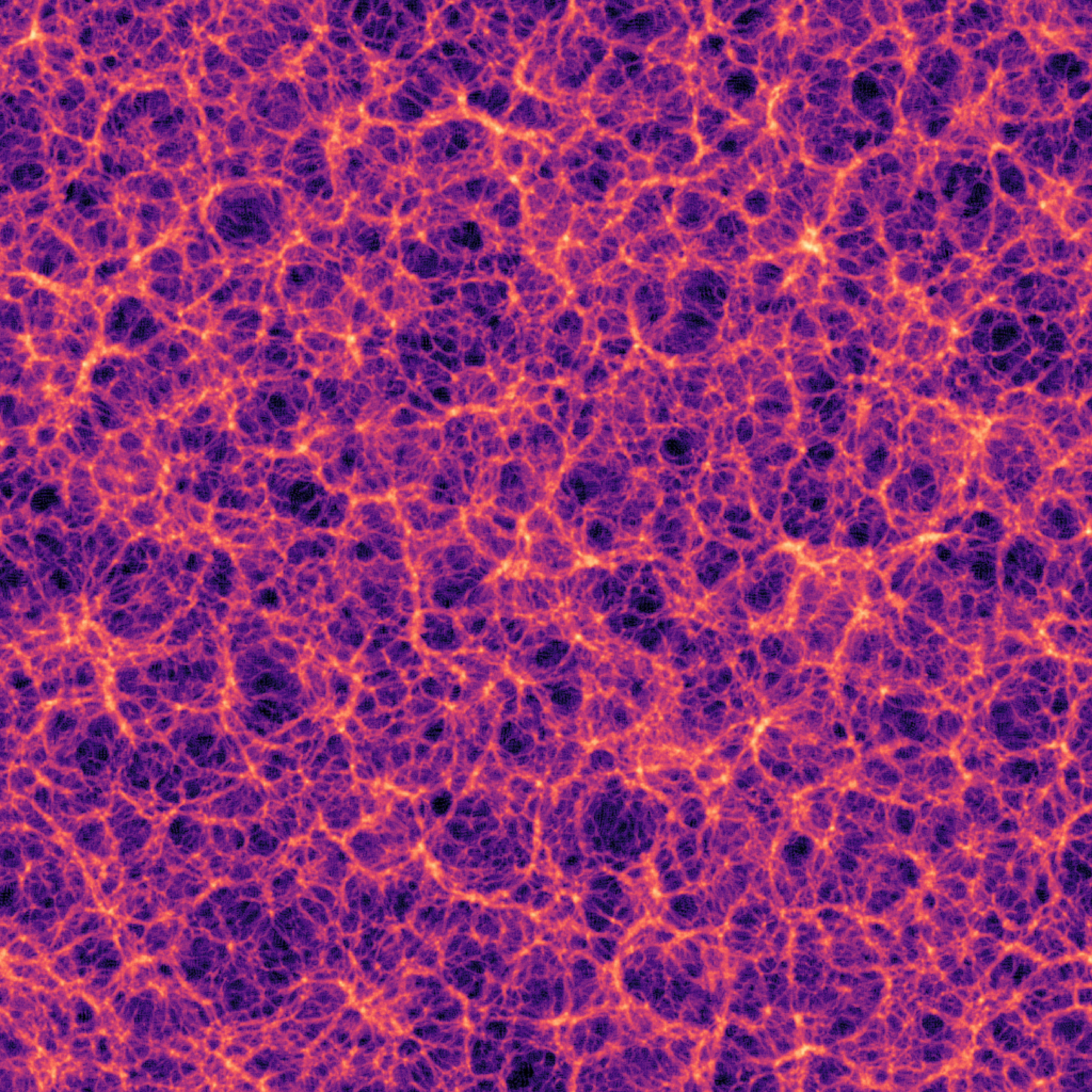
With Halo Exchange (Distributed)
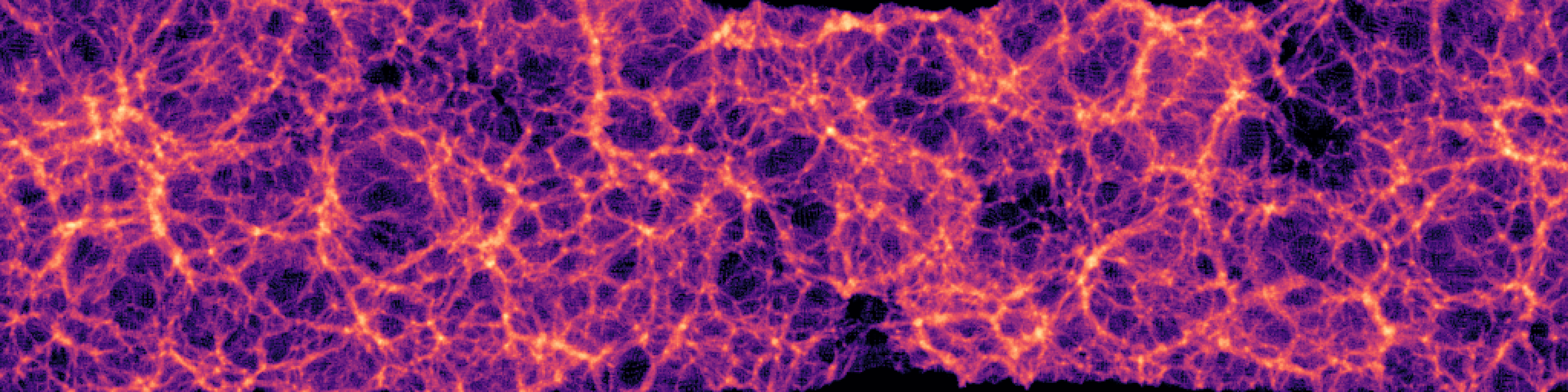
Why Halo Exchange Matters in Distributed Simulations


Without halo exchange, subdomain boundaries introduce visible artifacts in the final field.
This breaks the smoothness of the result — even when each local computation is correct.




What About the Gradients?
Backpropagating Through ODE Integration
Why Gradients Are Costly
To compute gradients through a simulation, we need to track:
- All intermediate positions: \(d_i\)
- All velocities: \(v_i\)
- And their tangent vectors for backpropagation
Even though each update is simple, autodiff requires storing the full history.
Example: Kick-Drift Integration
A typical update step looks like:
\[ \begin{aligned} d_{i+1} &= d_i + v_i \, \Delta t \\\\ v_{i+1} &= v_i + F(d_{i+1}) \, \Delta t \end{aligned} \]
Over many steps, the memory demand scales linearly — which becomes a bottleneck for large simulations.
Why This Is a Problem in Practice
Storing intermediate states for autodiff causes memory to scale linearly with the number of steps.
Example costs at full resolution:
- (1 Gpc/h)³, 10 steps → ~500 GB
- (2 Gpc/h)³, 10 steps → ~4.2 TB
This severely limits how many time steps or how large a volume we can afford — even with many GPUs.
Reverse Adjoint: Gradient Propagation Without Trajectory Storage (Preliminary)

Instead of storing the full trajectory…
We use the reverse adjoint method:
- Save only the final state
- Re-integrate backward in time to compute gradients
Forward Pass (Kick-Drift)
\[ \begin{aligned} d_{i+1} &= d_i + v_i \, \Delta t \\ v_{i+1} &= v_i + F(d_{i+1}) \, \Delta t \end{aligned} \]
Reverse Pass (Adjoint Method)
\[ \begin{aligned} v_i &= v_{i+1} - F(d_{i+1}) \, \Delta t \\ d_i &= d_{i+1} - v_i \, \Delta t \end{aligned} \]
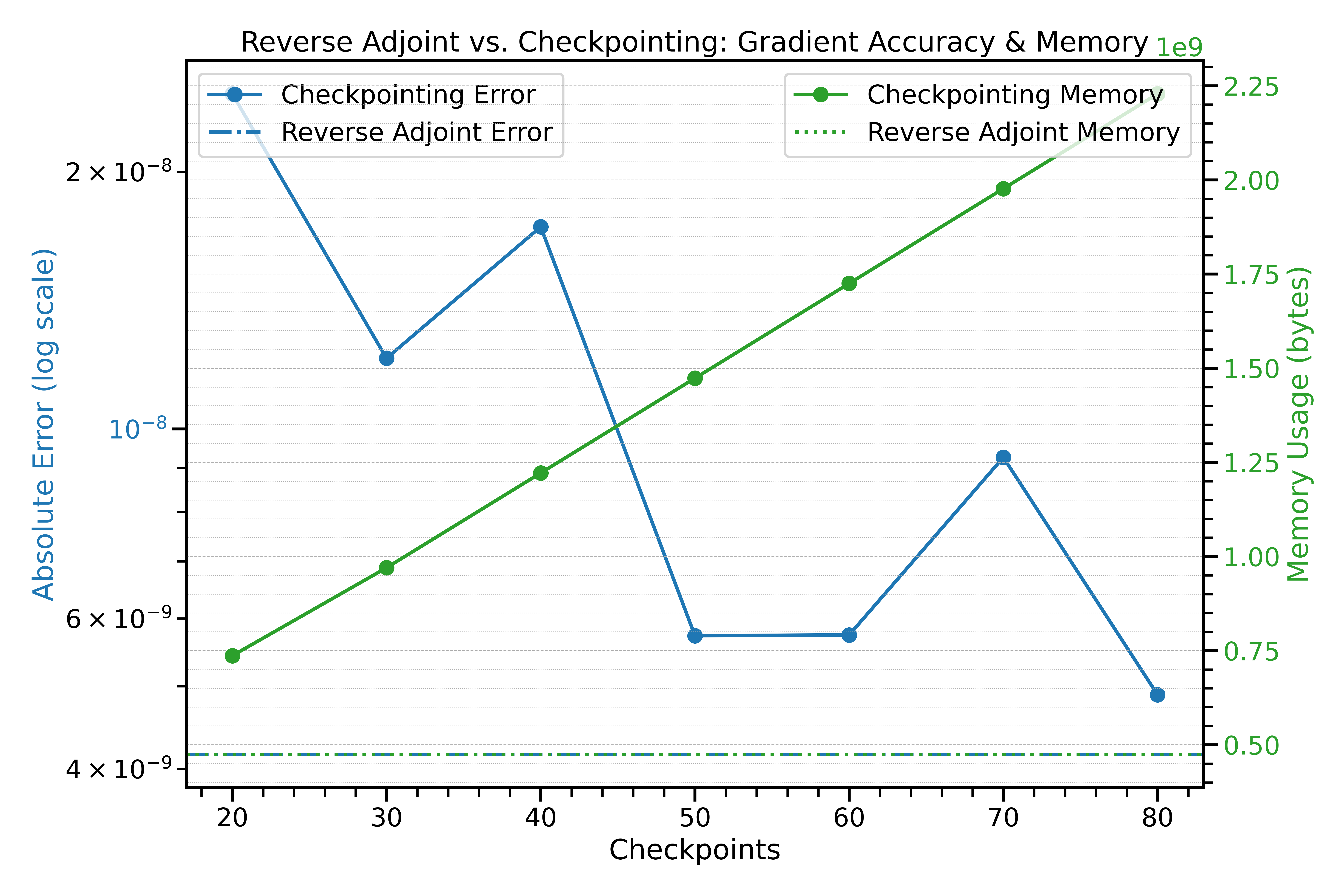
- Checkpointing saves intermediate simulation states periodically to reduce memory — but still grows with the number of steps.
- Reverse Adjoint recomputes on demand, keeping memory constant.
Reverse Adjoint Method
- Constant memory regardless of number of steps
- Requires a second simulation pass for gradient computation
- In a 10-step 1024³ Lightcone simulation, reverse adjoint uses 5× less memory than checkpointing (∼100 GB vs ∼500 GB)
Putting It All Together
JAXPM v0.1.5: Differentiable, Scalable Simulations
What JAXPM v0.1.5 Supports
Multi-GPU and Multi-Node simulation with distributed domain decomposition (Successfully ran 2048³ on 256 GPUs)
End-to-end differentiability, including force computation and interpolation
Compatible with a custom JAX compatible Reverse Adjoint solver for memory-efficient gradients
Supports full PM Lightcone Weak Lensing
Available on PyPI:
pip install jaxpmBuilt on top of
jaxdecompfor distributed 3D FFT
Current Capabilities & Road Ahead
What We’ve Achieved So Far
Built a scalable, differentiable N-body simulation pipeline (JAXPM)
Enables forward modeling and sampling in large cosmological volumes, paving the way toward full LSST-scale inference
Preliminary performance:
- ~20 s per 512×512×1024 simulation on 64×A100 GPUs
- <1 TB memory for full 10-step lightcone
- Stable gradients over 100-sample tests
For discussion
Using Scattering Transform to compress the data with SBI
Using Excplicit Inference for CMB r estimation
Extra slides
Approximating the Small Scales
Dynamic resultion grid
- We can use a dynamic resolution that automatically refines the grid to match the density regaions blabla
- very difficult to differentialte and slow to compute
Multigrid Methods
- Multigrid solves the Poisson equation efficiently by combining coarse and fine grids
- It’s still an approximation — it does not match the accuracy of solving on a uniformly fine grid
- At high fidelity, fine-grid solvers outperform multigrid in recovering small-scale structure — critical for unbiased inference
Bayesian Deep Learning Workshop , 2025